Updated 7 August 2023 (c) 2023
Evolution as a “theory”
One of the most common misconceptions in the public scientific arena is that evolution is “just a theory.” The Merriam-Webster dictionary lists several definitions for the word “theory” [Merriam2009]. The two that are the most relevant here are: (a) “a plausible or scientifically acceptable general principle or body of principles offered to explain phenomena, e.g., the wave theory of light” and (b) “a hypothesis assumed for the sake of argument or investigation; an unproved assumption.” In most scientific discourse, scientists use definition (a), while in the public arena (b) is more widely assumed. This distinction is the root of the widespread misunderstanding of the phrase “theory of evolution.” Evolution is most certainly not termed a “theory” because it is a sketchy conjecture that has never been seriously tested, but instead because it is a general principle with substantial explanatory power and falsifiability that has withstood very rigorous scrutiny over an extended period of time (see summary below).
Part of this misconception may be due to the fact that relatively few people are aware of the extensive scope of scientific research in general, and of research on evolution in particular. Along this line, a 2018 report by the U.S. National Science Board found that approximately 2.3 million scientific papers had been published in peer-reviewed journals worldwide in 2016, of which 15.3%, or approximately 350,000, are in biological sciences [NSB2018]. If one conservatively estimates that 10% of these directly touch on evolutionary science, and if one disregards papers in the other tabulated fields (chemistry, physics, geosciences, medical sciences, etc.) that may be relevant to evolution, then one still has at least 35,000 papers per year on some aspect of evolutionary science.
In any event, claims such as “evolution is just a theory,” as well as other arguments of a purely philosophical, social or historical nature, have no currency in the real world of scientific research one way or the other (see Postmodern). This is not merely an arbitrary convention among scientists; it is the consensus of decades and indeed centuries of experience in many fields of science. Instead, in the area of evolutionary science as well as in any other field, the fundamental touchstone is the process of proposing testable hypotheses and performing rigorous empirical analyses, using the best available technology and analytic tools. That is what we will address here.
What exactly is “evolution”?
The Merriam-Webster dictionary defines “evolution” as “a theory that the various types of animals and plants have their origin in other preexisting types and that the distinguishable differences are due to modifications in successive generations” [Merriam2009]. Even so, it should be emphasized, as briefly noted above, that evolution is a broad umbrella encompassing many fields of the life sciences, among them anatomy, anthropology, bacteriology, biochemistry, biology, biophysics, botany, ecology, entomology, epidemiology, genetics, geology, geophysics, molecular biology, microbiology, neurophysiology, paleontology, physiology, taxonomy, virology and zoology. For that matter, evolution touches numerous other fields outside the life sciences, including organic chemistry, physics, mathematics and computer science.
Thus it makes little sense to say one does or does not “believe in evolution” — one must be much more specific for any statement of this sort to have any crisp, empirically testable scientific substance. For that matter, scientists do not “believe” in evolution, nor do they say that evolution is “proven.” Instead, as noted above, they propose and perform rigorous tests for various hypotheses related to evolutionary science.
Here are just a few of the specific assertions normally included within the umbrella of evolutionary science:
- Life has existed and proliferated on Earth for at least 3.5 billion years, with most of the species that we can study as fossils arising in the past 600 million years. Mammals first appeared approximately 175 million years ago, and modern humans first appeared approximately 300,000 years ago.
- The vast majority of species that have existed on Earth are extinct (such as, for example, nearly all lines of dinosaur species).
- All living organisms on Earth today are descended from a small number of original ancestral species, quite possibly just one, since there is good evidence to support this hypothesis.
- The organization of present-day organisms into species, genus, family, order, class, phylum, kingdom and domain reflects actual biological common ancestry.
- Modern humans as a species arose, like every other species currently on Earth, from common ancestors. From anatomical, biochemical and DNA evidence, our closest living relatives are chimpanzees, followed by bonobos and gorillas.
- The principal driving forces for the formation and proliferation of species and features are the mostly random effects of mutations, transposons, gene duplication and hybridization, together with the mostly nonrandom effects of natural selection and environmental pressures.
- Similarities and differences in the DNA sequences between individual organisms and species reflect, in most cases, their actual genetic history.
So what are the principal lines of evidence that support these and other assertions? We will briefly summarize some of this evidence below. For more complete details, see the pages on the sites mentioned below and referenced books and articles in the bibliography.
Geologic evidence
One of the most compelling lines of evidence for evolution in general and the system of geologic dates in particular is the fact that the various geological eras, as identified by the fossils they contain, always appear in the same order (except in a few cases where there is clear evidence of overthrusting) and always yield the same geological dates, no matter where they are unearthed.
Geologic dates are derived from radiometric dating methods, which are based on known rates of radioactivity, a phenomenon that is rooted in fundamental laws of quantum physics and follows well-understood mathematical rules of exponential decay. The various radiometric dating schemes currently in use have been scrutinized and refined for over 70 years, and the latest high-tech equipment permits reliable results to be obtained even with microscopic samples. Further, radiometric dating is self-checking, because the data (after certain preliminary calculations are made) are fitted to a straight line by means of standard linear regression methods of statistics. Technical details on how these dates are calculated are given in Radiometric dating. For additional details on the reliability of radiometric methods, see Reliability.
Some young-earth creationists have claimed that scientists cannot know with any certainty what happened millions of years ago — one would need a “time machine” for this. But in a curious twist of irony, scientists really do have time machines, in the form of astronomical telescopes. This is because when an astronomer views, say, a Type 1A supernova explosion in the Pinwheel Galaxy (see photo), which is known by completely different and highly reliable lines of evidence to be approximately 21 million light-years away, these astronomers are witnessing an event that transpired approximately 21 million years ago. And such observations have confirmed in exquisite detail that the physical laws that governed the universe 21 million years ago, specifically including the laws of quantum mechanics and radioactive decay that are relevant to geologic dating, are experimentally indistinguishable to those laws observed in laboratories today.

In short, the geologic evidence for evolution, especially for the system of multimillion-year dates assigned to the various fossil layers and geologic epochs through the lifetime of planet Earth, is on extremely firm ground. To deny or blithely dismiss this conclusion, claiming instead that Earth was created a few thousand years ago, is tantamount to claiming that the tens of thousands of peer-reviewed radiometric age studies that have been published over the past 70 years, using numerous different dating techniques and approaches, are simultaneously in error, not just by a few percent here and there, but instead by factors of thousands and millions.
It is ironic that the one aspect of evolution that draws the most opposition, not only from the general public but also from the young-earth creationist community, namely the fact that Earth and its biosphere are vastly older than a few thousand years, is arguably the most experimentally solid of all.
Fossil evidence
The geologic record reveals hundreds of thousands of prehistoric species, preserved in stone, that have populated Earth over eons of geologic time, and researchers estimate that millions of species have existed on Earth over its 4.5-billion-year lifetime.
Creationists have repeatedly emphasized “gaps” in the fossil record, seeking to cast doubt on this conclusion. It is undeniably true that there are gaps (no responsible geologist has ever claimed otherwise), but it is also true that many of the gaps once highlighted by creationists have subsequently been filled. With regard to gaps, it must be kept in mind that the discovery of a fossil is a highly fortuitous event. Virtually all biological organisms that have ever lived were either eaten by predators or otherwise destroyed soon after death, leaving no trace. Most that persisted in some form (e.g., as skeletons) were later destroyed by chemical effects, or were part of a geological layer that subsequently disappeared into Earth’s molten mantle, or are far beneath Earth’s surface and will never be found by humans. Also, it is not always clear what species is descended from which, just by examination of fossils — in many cases, features arose from some previous biologically useful purposes before being adapted for a later purpose. For example, feathers arose for warmth long before they were used for flight, and air sacs were used for flotation long before they were used for breathing [Shubin2020].
In spite of these challenges, many highly significant transitional fossils have been found in recent years, generously filling “gaps” that once were claimed by creationists and others to be unbridgeable. Here are just a handful of examples:
- Icthyosaurs. Creationists have long listed ichthyosaurs, namely dolphin-like reptiles that lived in the ocean during the age of dinosaurs, as a leading counter-example to evolutionary theory. Ichthyosaurs are hypothesized to have evolved from earlier terrestrial creatures that subsequently re-entered the water, yet until recently no transitional fossils were known. But in November 2014, a team of researchers from the U.S., Italy and China announced that they have discovered a perfect intermediate fossil, a 50 cm-long specimen that lived 248 million years ago. Unlike ichthyosaurs, this species had large flippers to facilitate walking on land like a seal [Feltman2014].
- Marine-land mammal transition. Both creationists and intelligent design writers have questioned whether scientists would ever discover fossils linking the hypothesized transition between land mammals and marine mammals. Creationist Duane Gish, for example, has highlighted his presentations on this topic by displaying a cartoon of “Bossie” the cow, “Blowhole” the whale and a ridiculous intermediate creature between a cow and whale [Gish1985, pg. 78-79; Prothero2007, pg. 318]. Yet at least 30 distinct intermediate fossil species are now known within this “gap,” with exactly the expected combination of terrestrial and aquatic features. For a summary of these intermediate forms, see Fossils.
- Fish-tetrapod transition. In another striking recent development, scientists discovered a long-sought transition between bony fish and tetrapods (four-legged animals including reptiles, birds, mammals, humans). Named “Tiktaalik,” it was discovered on a remote island in the Canadian Arctic Ocean, in sedimentary rock 375 million years old. Although this creature had many features of fish, it also had several traits characteristic of early tetrapods, including fins that contained bones, forming a limb-like appendage that the animal could use for locomotion on land [NAS2008, pg. 1-3; Prothero2007, pg. 228-229]. A fascinating account of this discovery, by the researcher himself, is given in [Shubin2008, pg. 1-27]. Here is a sketch of how Tiktaalik fits into known intermediate forms:
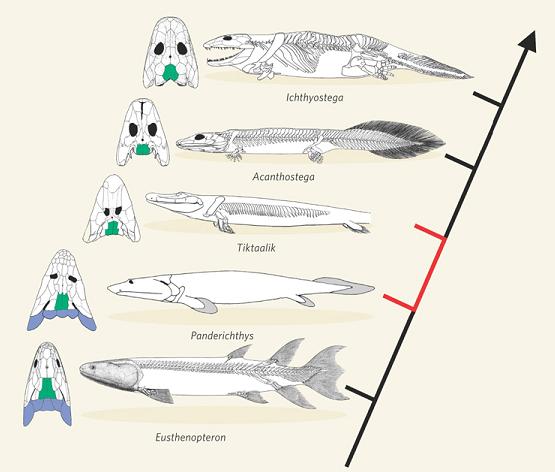
- Elephants. Twenty-two distinct species of elephants have now been identified during just the past six million years, generously filling the “gap” between ancient forms and four modern species (the three currently existing elephant species and the recently extinct wooly mammoth). One recently compiled family tree of these species is shown at Fossils.
- Dinosaur-bird transition. Numerous fossils have been found confirming a hypothesis originally made by Thomas Huxley that birds are modern descendants of dinosaur-like creatures. In 2008 Chinese paleontologist Xu Xing and his colleagues documented what appears to be the simplest type of “protofeather,” which was seen in a therizinosaur fossil. Another remarkable development in this area was the 2010 discovery by Xu Xing’s team of a 160-million-year-old dinosaur fossil in northwestern China that had a bird-like keel-shaped chest and a long beak, yet also had three toes on each leg like a dinosaur [Perlman2010]. As a result of these and other findings, the overwhelming majority of paleontologists are now convinced that modern birds are descendants of certain dinosaurs. A nice overview of recent research in this field is given in a Quanta article by Emily Singer [Singer2015].
For additional details on these and several other examples, see Fossils and Prehuman fossils.
Novel structures
Creationist and intelligent design writers have long insisted that “undirected” evolution cannot possibly create any new, useful biological structures or features. Yet researchers have documented many examples of exactly this happening. Here are a few examples:
- The Hall-Hartl E. coli experiment. In a 1974 paper Barry Hall and Daniel Hartl identified a gene in the bacterium E. coli that is responsible for metabolizing lactose, using a complicated three-part process. They removed this gene, and then permitted the bacteria to multiply in a stressed environment containing lactose. Within 24 hours the bacteria had evolved a capability to utilize lactose, by means of a similar but distinct three-part biochemical pathway, involving two mutated genes [Hall1974; Miller1999, pg. 145-147].
- Lenski’s long-running E. coli experiment. Biologist Richard Lenski and his colleagues have been conducting a long-running experiment on bacterial evolution that began in 1988. Starting with 12 flasks of E. coli bacteria, identical except for some neutral markers, and then each day inserting 1/100 of the flask’s liquid (which contained glucose and citrate, among other materials) into a new flask. In this way they followed the course of these bacteria for 45,000 generations. As the generations continued, each of the 12 lines grew progressively better at processing glucose, although each took a different trajectory. Later in the experiment, shortly after generation 33,000, the average population of one of the lines shot up by a factor of six above the others. The investigators found that this line had developed the ability to utilize citrate, which bacteria normally cannot use, by means of a remarkable combination of two distinct mutations [Lenski1994; Dawkins2009, pg. 116-132].
In a new result from Lenski’s long-running experiment, in June 2015 Lenski’s team found that that a second strain of E. coli has arisen within the same flask as the citrate-eating strain. This second strain, although lacking the ability to utilize citrate, has evolved the ability to utilize some by-products of the first strain. Thus the two strains together have formed a cross-feeding ecosystem, all within a flask [Turner2015].
- Lenski’s 2012 virus/E. coli experiment. In a separate experimental study, a research team led by Lenski demonstrated how colonies of viruses were able to evolve a new trait in as little as 15 days. The researchers studied a virus, known as “lambda,” which infects only the bacterium E. coli. They engineered a strain of E. coli that had almost none of the molecules that this virus normally attaches to, then released them into the virus colony. In 24 of 96 separate experimental lines, the viruses evolved a strain that enabled them able to attach to E. coli, using a new molecule (a channel in E. coli known as “OmpF”) that they had never before been observed to utilize. All of the successful runs utilized essentially the same set of four distinct mutations [Zimmer2012].
- Japanese nylon-eating bacteria. Japanese biologists recently discovered a bacterial species that thrives in nylon waste. It turns out that these bacteria had undergone a “frame shift” mutation, where an extra base pair had been inserted into the bacteria’s DNA. This mutation significantly changed the bacteria’s biology, since a long series of amino acids were altered, but by remarkable chance this alteration endowed the bacteria with the facility to metabolize nylon, albeit not very efficiently [Negoro1994].
- The Milano mutation. Scientists recently discovered that certain persons in an Italian community, all descended from a single individual several generations back, possess a genetic mutation that increases “good” cholesterol and provides an effective antioxidant, thus resulting in measurably improved cardiovascular health [Krotz2002; Musgrave2003].
- Antibiotic-resistant diseases. Perhaps the best-known examples are the recent evolution of new strains of tuberculosis that are resistant to all known anti-TB drugs. By analyzing DNA sequences, researchers have identified at least six different families of tuberculosis, at least one of which appears to be evolving on an unexpected and potentially very dangerous path [Lehrman2013]. Another example is drug-resistant strains of HIV that in many cases evolve within the body of a single patient [Coyne2009, pg. 130-131; Mason2009].
- Tibetan high-altitude genes. In 2010, researchers at the University of Utah and Qinghai University in China have found that natives of the Tibetan highlands have evolved ten unique genes that permit them to live well at very high altitudes. Because of these genes, Tibetans have more efficient metabolisms, do not overproduce red blood cells in response to thin air, and have higher levels of nitric oxide, which helps get oxygen to tissue [SD2010b]. A even more recent study found a total of 30 genes that were distinct in the Tibetan population, and concluded that this change constitutes the fastest documented case of human evolution [Wade2010b].
- Hawaiian crickets. In the 1800s, a species of cricket was introduced to the Hawaiian Islands, where they became quite common in grassy areas. Males attract mates with their chirps, and females select males based on their songs. However, unlike their counterparts in other Pacific islands, the Hawaiian crickets have a fearsome predator — dive-bombing flies that target chirping crickets, then implant their larvae in them. In the 1990s, researchers noted that a field in Kauai that previously was the home to many crickets now seemed silent. However, a nighttime search found that in fact there were lots of crickets there, but very few of the males now chirped — in just five years, or roughly 20 generations, a mutation had arisen in a single gene that inhibited many of the males from chirping [Zuk2013, pg. 81-82].
- Evolution of the coronavirus. The Covid-19 pandemic of 2020-2023, which as of January 2023 has claimed the lives of over 18,000,000 worldwide, including over 1,000,000 Americans, has underscored the reality of evolutionary novelty in a way that affects every person on the planet. The rise of the delta and omicron variants of the virus (the latest is “XBB.1.5”), which are significantly more transmissible than earlier variants, is a particularly striking case of “evolution before our eyes” [Achenbach2021; Adam2022; Schumaker2021; Zimmer2023]. No one “designed” these deadly virus variants.
Speciation
Creationists and others have also claimed that the splitting of a species into two or more species has never been observed in nature. But biologists can cite numerous examples of present-day species that appear to be in the process of splitting and species that have split very recently in geologic history [Coyne2009, pg. 5-8, 168-189]. Some examples include:
- Galapagos finches. Charles Darwin, who first studied the finch populations on the Galapagos Islands, believed that speciation required hundreds or even thousands of generations. Yet a remarkable instance of speciation has just been observed among these finches. In 1981 researchers observed a single male finch, normally residing on either Espanola or Gardner Island, on the Island Daphne Major. To their surprise, within two generations a hybrid species had taken hold, exclusively breeding only with other finches descended from the original male [Cepelewicz2017].
- Salamanders. Ensatina eschscholtzi is a lungless salamander that ranges along the Pacific Coast from Canada to Mexico. Within this population, seven “subspecies” have been recognized in a ring around the Central Valley of California. About 35 miles southeast of Mount Palomar, near Cuyamaca State Park, these subspecies meet and fail to interbreed — in other words, the two subspecies in this area are different species by the usual definition of the term [Wake1986; Wake2001].
- Killer whales. In 2010 British researchers announced that two different types of killer whales (orcas) had been identified in the waters near the U.K. Each type differs somewhat in size and diet, and genetic analysis indicates that the two types belong to different populations that are in the process of becoming different species [Bourton2010]. More recently (April 2010), researchers confirmed that in fact there are three distinct populations of orcas whose DNA is sufficiently distinct that the three groups could be considered distinct species [Wade2010].
- Nicaraguan fish. In a 2010 study, researchers have found that fish in a remote crater lake in Nicaragua are rapidly splitting into separate species. In just 100 generations (over roughly 100 years), one group has developed very fat lips. The fat-lipped fish occupy a different ecological niche from the thin-lipped variety, even though they live together in the same lake — the fat-lipped group prefers insects, while the thin-lipped group prefers snails. The fish do not mate with members of the other group in the wild, although lab experiments show that they still can interbreed. The fact that they do not intermix in the wild indicates that they are in the process of becoming separate species [Coghlan2010a].
- Drosophila flies in Hawaii. Similar “sister species” of Drosophila flies have been found in Hawaii. In other words, these are closely related corresponding species that are found on different islands in the Hawaiian Island chain, which evidently split in recent geologic history. In this case, the dates of the speciation events have been determined by analyzing the flies’ DNA. As scientists had predicted, the oldest species have been found on the oldest islands [Coyne2009, pg. 181].
- Malaria-bearing mosquitos. A 2010 study of two strains of the Anopheles gambiae mosquito, which are the principal carriers of malaria in sub-Saharan Africa, has verified that these strains have diverged so much genetically that even though they still look exactly the same, they are now, in effect, two distinct species (e.g., because they no longer interbreed) [Nordqvist2010]. As the researchers who performed this study note, “From our new studies, we can see that mosquitoes are evolving more quickly than we thought and that unfortunately, strategies that might work against one strain of mosquito might not be effective against another. It’s important to identify and monitor these hidden genetic changes in mosquitoes if we are to succeed in bringing malaria under control by targeting mosquitoes.” [Neafsey2010].
- Stickleback fish in Alaska. In 1990, biologist Michael Bell found to his surprise that marine stickleback fish had recolonized Loberg Lake in Alaska, after being exterminated in 1982. What’s more, a few of these fish had developed features more typical of fresh water stickleback species, such as loss of body armor and changes in feeding structures in the throat. He and his team have returned to the lake each year since then and have documented a steady increase in the percentage of fresh-water-typical features. For instance, Bell and his graduate student Windsor Aguirre have observed that the lake fish population has changed from mostly 30 or more armor plates per side to mostly between five and eight plates per side. As Bell notes, “it has become clear that populations can evolve substantially on contemporary time scales and that the magnitude of evolutionary divergence between ancestral and descendant populations can be comparable to differences among related species” [Bell2004; LePage2011]. Here are photos of a typical marine stickleback (top) and two typical Loberg Lake stickleback (bottom), courtesy Michael Bell:

- Stickleback fish in Switzerland. In yet another example of speciation of stickleback fish, researchers at the University of Bern have found that a population of these fish in Switzerland’s Lake Constance, which was only introduced roughly 150 years ago, appears to have split into two species — one lives in the main lake, and the other lives in streams flowing into the lake. The lake fish are generally larger, and have longer spines and tougher armor. The researchers confirmed that it was speciation, not just lifestyle, by noting clear genetic differences between the two populations [LePage2016].
DNA evidence
Direct analyses of DNA sequences (either whole or parts) have been made possible by the recent dramatic advances in DNA sequencing technology. For example, the cost of producing a complete genome of a human has declined from roughly three billion dollars in 2003, when the first human genome was sequenced, to less than $300 today, and the price will likely fall to only $100 by 2023 [Vance2014; Fikes2017; [Coldewey2022]. This astonishing drop, by roughly a factor of ten million over a 17-year period (roughly three times as fast as Moore’s Law of semiconductor technology), has enabled numerous advances in biomedicine in general, and in evolutionary biology in particular.
Such advances, which were completely unforeseen by biologists of earlier eras, have dramatically confirmed many of the basic tenets of evolutionary science. Here are just a few examples:
- Comparisons of beta globin among species. One example of DNA-type data is comparisons of the 146-unit amino acid sequences of beta globin (a component of hemoglobin in blood) among various species of animals. Amino acids are coded directly by triplets of DNA letters, and thus the study of amino acid sequences is very close to the study of DNA sequences themselves. As it turns out, human beta globin is identical to that of chimpanzees, differs in only one location from that of gorillas, yet is increasingly distinct from that in red foxes, polar bears, horses, rats, chicken and salmon. For details, see the table at DNA.
- Mutations. The picture is the same if we consider the pattern of mutations between closely related species. For example, the gene that, when mutated, results in cystic fibrosis in humans is nearly identical to the corresponding gene in chimpanzees, but is progressively less similar to the corresponding gene in orangutans, baboons, marmosets, lemurs, mice, chicken and puffer fish [NAS2008, pg. 30]. As yet another example, cytochrome c, which is essential for cell respiration, differs only in one location out of 104 between humans and rhesus monkeys. Comparing humans and horses, there as 12 differences; comparing rhesus monkeys with horses, there are 11 differences. Evidently the single difference between humans and rhesus monkeys occurred after our hominid ancestors split from the lineage that led to present-day monkeys [Ayala2007, pg. 128-129].
- GULO gene for Vitamin C. The “GULO” gene is an essential part of the machinery that makes Vitamin C in most animals. Unfortunately, humans and other great apes lack a functioning copy of this gene — our copy is a highly mutated fragment, classified as a relic gene or pseudogene. Scurvy, that scourge of British seamen, Mormon pioneers crossing the Great Plains, and millions in poor regions worldwide even today, results when humans don’t get enough Vitamin C in their diets to compensate for the lack of a functioning GULO gene. Interestingly, although the GULO pseudogene is highly mutated and utterly useless, the human and chimp versions of this gene are 98% identical. Evidently a common ancestor of humans and chimps adopted a diet rich in fruits and vegetables, and thus a chance mutation that disabled Vitamin C production was no longer a fatal one and was passed on to posterity [Fairbanks2007, pg. 53-55; Coyne2009, pg. 67-69].
Along this line, the mutated and useless human GULO gene is an effective refutation of the claim by many creationist and intelligent design writers that each individual species was meticulously designed. After all, it would be strange indeed to insist that a Supreme Being “designed” humans with such a glaring defect, and it would be downright blasphemous to insist that this Being then copied this defect to several other primate species. Many more examples of this sort could be cited — see Design.
- Transposons. Transposons or “jumping genes” are sections of DNA that have been “copied” from one part of an organism’s genome and “pasted” seemingly at random in other locations. The human genome, for example, has over four million individual transposons, organized in over 800 families [Mills2007]. In most cases transposons do no harm, because they “land” in an unused section of DNA, but because they are inherited they serve as excellent markers for genetic studies. Indeed, transposons have been used to classify a large number of vertebrate species into a family tree, with a result that is virtually identical to what biologists had earlier reckoned based only physical features and biological functions [Rogers2011, pg. 25-31, 86-92]. As just one example, consider the following table, where columns labeled ABCDE denote five blocks of transposons, and x and o denote that the block is present or absent in the genome [Rogers2011, pg. 89].
Transposon blocks Species A B C D E /-------- Human o x x x x /--------- Bonobo x x x x x / \-------- Chimp x x x x x /----------- Gorilla o o x x x -----|----------- Orangutan o o o x x \----------- Gibbon o o o o oIt is clear from these data that our closest primate relatives are chimpanzees and bonobos. As another example, here is a classification of four cetaceans (ocean mammals) based on transposon data [Rogers2011, pg. 27]:
Transposon blocks Species A B C D E F G H I J K L M N O P /------ Bottlenose dolphin x x x x x x x x x x x x x x x x /\------ Narwhal whale x x x x x x x x x x x x x x x x ---|------- Sperm whale x x x x x o o o o o o o o o o o \------- Humpback whale x x o o o o o o o o o o o o o oOther examples could be listed, encompassing an even broader range of species [Rogers2011, pg. 25-31, 86-92]. For other, even more compelling details and additional discussion, see DNA.
- Chromosome fusion in humans. Biologists noted long ago that humans have only 23 pairs of chromosomes, whereas other great apes — chimpanzees, bonobos, gorillas and orangutans — have 24. Thus they were led to conjecture that two of the human chromosomes have fused since the split between ancestral human and ape lineages. This hypothesis gained credence in 1982, when scientists found that chromosomes from humans, chimpanzees, gorillas and orangutans are highly similar and can be aligned with one another, with human chromosome #2 corresponding to the slightly overlapped union of ape chromosomes 2A and 2B (graphic credit: Memorial University of Newfoundland, Canada):
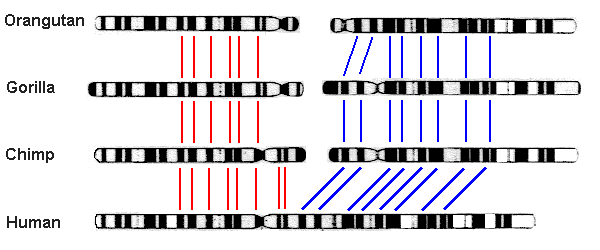
The final confirmation came in 1991 from a detailed analysis of human DNA, which found two complementary telomeres (repeated sequences of a certain DNA string that appear at the end of a chromosome) spanning the exact spot of union [Fairbanks2007, pg. 20-27; Fairbanks2012, pg. 135-139]:
Fusion site | ... TTAGGGG TTAGGG TTAG CTAA CCCTAA CCCTAA ... ... AATCCCC AATCCC AATC GATT GGGATT GGGATT ... |(Note that the second row is almost exactly a reversal of the first, pivoted about the fusion site.) Numerous other similar examples are given in Fairbank’s book Evolution: The Human Effect and Why It Matters [Fairbanks2012].
- The genetic code and evolution. Although the “genetic code,” namely the system of assigning 3-letter DNA sequences to one of the 20 amino acids employed in biology, is universal over almost all the biological kingdom, there are a few exceptions. For example, mitochondria, the little “islands” within a cell that generate most of the cell’s chemical energy, employ a slightly different version. In total, scientists have by now identified 34 different codes in the biological kingdom. Yet, as biologist Kenneth Miller observes, these variant genetic codes are all neatly arranged in a hierarchical pattern, like variant dialects of English, which pattern is compelling evidence for their common ancestry [Miller2001; Zimmer2013].
- DNA data and the human-chimpanzee split. Researchers are also combining analyses of DNA sequences with paleontological (fossil) data, resulting in more precise determinations of various branches in the tree of life. For example, a study published in November 2010 that combined both paleontological and molecular data established that divergence of humans and chimpanzees very likely took place roughly eight million years ago, instead of five to six million years as generally believed until recently [SD2010d; Wilkinson2010]. In November 2012, a new study observed a rate of 36 new mutations per human generation (half the earlier estimate), which was obtained from whole-genome DNA sequencing of 78 persons and their biological parents. As a result of such analyses, the current consensus is that the human-chimpanzee split occurred between 7 and 13 million years ago [Brahic2012].
- DNA and phylogenetics. Researchers are analyzing DNA of groups of existing species to reconstruct their “family tree.” Soon much of evolutionary history will be deducible purely from this type of automatic computer-based analysis. For example, in May 2010 a researcher announced, on the basis of a very carefully performed statistical analysis, that the hypothesis of a “universal common ancestor” hypothesis (a conjecture, dating back to Charles Darwin, that all life arose from a single common ancestral species) is at least 102860 times more likely to have produced the modern-day protein sequences that we observe in living organisms, compared to the next most probable scenario that involves multiple original ancestral species [Harmon2010; Theobald2010].
For additional details on these and several other examples, see DNA.
Conclusions
Any one of these lines of evidence constitutes very strong evidence for the basic tenets of evolutionary science; together they are as unassailable as any theory of science. It might be mentioned that several of these lines of evidence, notably radiometric dating, comparisons of protein sequences and analysis of DNA, were completely unforeseen at the time of Charles Darwin. Doubtless he would have been astounded that his theory could be rigorously analyzed and tested by such powerful tools — tools that continue to improve at a rapid rate, as in the case of DNA sequencing.
In summary, evolution is indeed a “theory” in the strict scientific sense, namely as an umbrella for numerous specific scientific assertions that have been studied in great empirical detail and have survived many rigorous tests. Thus those who promote the line that evolution is “only a theory” (implying a sketchy hypothesis that has never been rigorously tested), or who otherwise blithely dismiss the huge corpus of evidence for evolution either: (a) are completely uninformed as to the true nature of the scientific enterprise; (b) are oblivious to the hundreds of thousands of published research studies in the field; or (c) are being deliberately disingenuous to their audience. Rigorous scientific research will continue, and doubtless our understanding of certain specific details of evolution will change in the coming years, but these changes will come from professionally conducted, peer-reviewed studies.